Protein-coding gene
| ELF5 |
|---|
 |
| Available structures |
|---|
| PDB | Ortholog search: PDBe RCSB |
|---|
|
|
| Identifiers |
|---|
| Aliases | ELF5, ESE2, E74 like ETS transcription factor 5 |
|---|
| External IDs | OMIM: 605169; MGI: 1335079; HomoloGene: 7702; GeneCards: ELF5; OMA:ELF5 - orthologs |
|---|
| Gene location (Human) |
|---|
 | | Chr. | Chromosome 11 (human)[1] |
|---|
| | Band | 11p13 | Start | 34,478,791 bp[1] |
|---|
| End | 34,525,193 bp[1] |
|---|
|
| Gene location (Mouse) |
|---|
 | | Chr. | Chromosome 2 (mouse)[2] |
|---|
| | Band | 2|2 E2 | Start | 103,242,033 bp[2] |
|---|
| End | 103,281,334 bp[2] |
|---|
|
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
| Top expressed in | - parotid gland
- amniotic fluid
- seminal vesicula
- epithelium of lactiferous gland
- lactiferous duct
- minor salivary glands
- olfactory zone of nasal mucosa
- mucosa of paranasal sinus
- buccal mucosa cell
- palpebral conjunctiva
|
| | Top expressed in | - seminal vesicula
- parotid gland
- lactiferous gland
- olfactory epithelium
- left lung lobe
- submandibular gland
- lacrimal gland
- medullary collecting duct
- esophagus
- lip
|
| | More reference expression data |
|
|---|
| BioGPS | 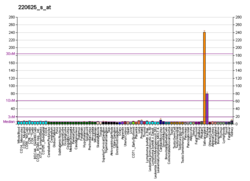
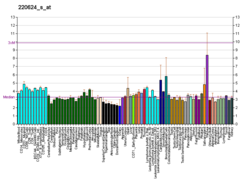 | | More reference expression data |
|
|---|
|
| Gene ontology |
|---|
| Molecular function | - DNA-binding transcription activator activity, RNA polymerase II-specific
- DNA binding
- DNA-binding transcription factor activity
- sequence-specific DNA binding
- RNA polymerase II transcription regulatory region sequence-specific DNA binding
- DNA-binding transcription factor activity, RNA polymerase II-specific
| | Cellular component | | | Biological process | - regulation of transcription by RNA polymerase II
- ectodermal cell fate commitment
- negative regulation of cell differentiation
- cell differentiation
- ectoderm development
- cell population proliferation
- regulation of transcription, DNA-templated
- somatic stem cell population maintenance
- transcription, DNA-templated
- positive regulation of transcription by RNA polymerase II
- mammary gland epithelial cell differentiation
- transcription by RNA polymerase II
| | Sources:Amigo / QuickGO |
|
| Orthologs |
|---|
| Species | Human | Mouse |
|---|
| Entrez | | |
|---|
| Ensembl | | |
|---|
| UniProt | | |
|---|
| RefSeq (mRNA) | |
|---|
NM_198381
NM_001243080
NM_001243081
NM_001422 |
| |
|---|
| RefSeq (protein) | |
|---|
NP_001230009
NP_001230010
NP_001413
NP_938195 |
| |
|---|
| Location (UCSC) | Chr 11: 34.48 – 34.53 Mb | Chr 2: 103.24 – 103.28 Mb |
|---|
| PubMed search | [3] | [4] |
|---|
|
| Wikidata |
| View/Edit Human | View/Edit Mouse |
|
E74-like factor 5 (ets domain transcription factor), is a gene found in both mice and humans.[5] In humans it is also called ESE2.
Function
The protein encoded by this gene is a member of an epithelium-specific subclass of the ETS transcription factor family. In addition to its role in regulating the later stages of terminal differentiation of keratinocytes, it appears to regulate a number of epithelium-specific genes found in tissues containing glandular epithelium such as salivary gland and prostate. It has very low affinity to DNA due to its negative regulatory domain at the amino terminus. Two alternatively spliced transcript variants encoding different isoforms have been described for this gene.[5]
Relevance to Disease
A role in breast or prostate cancer is known.[6] There are preliminary reports that the C allele genetic variant of rs61882275 near chromosome location 11.13 with variable incidence in human populations[7] is associated with severe COVID-19.[8] Further investigation by others suggested the association was caused by a tissue-specific effect on ELF5 expression in lung endothelium by this allele which results in a more than 4-fold higher risk of severe COVID-19 in those with SARS-CoV2 infection.[9]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000135374 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000027186 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b "Entrez Gene: ELF5 E74-like factor 5 (ets domain transcription factor)".
- ^ Piggin CL, Roden DL, Gallego-Ortega D, Lee HJ, Oakes SR, Ormandy CJ (7 January 2016). "ELF5 isoform expression is tissue-specific and significantly altered in cancer". Breast Cancer Research. 18 (1). 4. doi:10.1186/s13058-015-0666-0. PMC 4704400. PMID 26738740.
- ^ "dbSNP Short Genetic Variations: rs61882275". Retrieved January 21, 2022.
- ^ Kousathanas A, et al. (8 September 2021). "Whole genome sequencing identifies multiple loci for critical illness caused by COVID-19". medRxiv 10.1101/2021.09.02.21262965.
- ^ Pietzner M, et al. (19 January 2022). "ELF5 is a respiratory epithelial cell-specific risk gene for severe COVID-19". medRxiv 10.1101/2022.01.17.22269283.
Further reading
- Sharrocks AD, Brown AL, Ling Y, Yates PR (Dec 1997). "The ETS-domain transcription factor family". The International Journal of Biochemistry & Cell Biology. 29 (12): 1371–87. doi:10.1016/S1357-2725(97)00086-1. PMID 9570133.
- Zhou J, Ng AY, Tymms MJ, Jermiin LS, Seth AK, Thomas RS, Kola I (Nov 1998). "A novel transcription factor, ELF5, belongs to the ELF subfamily of ETS genes and maps to human chromosome 11p13-15, a region subject to LOH and rearrangement in human carcinoma cell lines". Oncogene. 17 (21): 2719–32. doi:10.1038/sj.onc.1202198. PMID 9840936.
- Oettgen P, Kas K, Dube A, Gu X, Grall F, Thamrongsak U, Akbarali Y, Finger E, Boltax J, Endress G, Munger K, Kunsch C, Libermann TA (Oct 1999). "Characterization of ESE-2, a novel ESE-1-related Ets transcription factor that is restricted to glandular epithelium and differentiated keratinocytes". The Journal of Biological Chemistry. 274 (41): 29439–52. doi:10.1074/jbc.274.41.29439. PMID 10506207.
- Lapinskas EJ, Palmer J, Ricardo S, Hertzog PJ, Hammacher A, Pritchard MA (Dec 2004). "A major site of expression of the ets transcription factor Elf5 is epithelia of exocrine glands". Histochemistry and Cell Biology. 122 (6): 521–6. doi:10.1007/s00418-004-0713-x. PMID 15655699. S2CID 28936823.
- Yaniw D, Hu J (Jun 2005). "Epithelium-specific ets transcription factor 2 upregulates cytokeratin 18 expression in pulmonary epithelial cells through an interaction with cytokeratin 18 intron 1". Cell Research. 15 (6): 423–9. doi:10.1038/sj.cr.7290310. PMC 4494830. PMID 15987600.
- Tummala R, Sinha S (Mar 2006). "Differentiation-specific transcriptional regulation of the ESE-2 gene by a novel keratinocyte-restricted factor". Journal of Cellular Biochemistry. 97 (4): 766–81. doi:10.1002/jcb.20685. PMID 16229011. S2CID 46125527.
- Choi YS, Sinha S (Sep 2006). "Determination of the consensus DNA-binding sequence and a transcriptional activation domain for ESE-2". The Biochemical Journal. 398 (3): 497–507. doi:10.1042/BJ20060375. PMC 1559455. PMID 16704374.
- Lim J, Hao T, Shaw C, Patel AJ, Szabó G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabási AL, Vidal M, Zoghbi HY (May 2006). "A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration". Cell. 125 (4): 801–14. doi:10.1016/j.cell.2006.03.032. PMID 16713569. S2CID 13709685.
External links
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
|
|---|
(1) Basic domains |
|---|
| (1.1) Basic leucine zipper (bZIP) | |
|---|
| (1.2) Basic helix-loop-helix (bHLH) | | Group A | |
|---|
| Group B | |
|---|
Group C
bHLH-PAS | |
|---|
| Group D | |
|---|
| Group E | |
|---|
Group F
bHLH-COE | |
|---|
|
|---|
| (1.3) bHLH-ZIP | |
|---|
| (1.4) NF-1 | |
|---|
| (1.5) RF-X | |
|---|
| (1.6) Basic helix-span-helix (bHSH) | |
|---|
|
|
(2) Zinc finger DNA-binding domains |
|---|
| (2.1) Nuclear receptor (Cys4) | | subfamily 1 | |
|---|
| subfamily 2 | |
|---|
| subfamily 3 | |
|---|
| subfamily 4 | |
|---|
| subfamily 5 | |
|---|
| subfamily 6 | |
|---|
| subfamily 0 | |
|---|
|
|---|
| (2.2) Other Cys4 | |
|---|
| (2.3) Cys2His2 | |
|---|
| (2.4) Cys6 | |
|---|
| (2.5) Alternating composition | |
|---|
| (2.6) WRKY | |
|---|
|
|
|
(4) β-Scaffold factors with minor groove contacts |
|---|
|
|
(0) Other transcription factors |
|---|
|
|
see also transcription factor/coregulator deficiencies |






















